May 06, 2016
I’ve been trying to get LaTeX working for a while. Turns out this is the magic recipe, though the unnumbered equations still don’t work quite right. I don’t foresee myself using them, though.
Let’s test some inline math $x$, $y$, $x_1$, $y_1$.
Now inline math with a special character: $|\psi\rangle$, $x’$, $x^*$.
Test display math:
$$ |\psi_1\rangle = a|0\rangle + b|1\rangle $$
Is it ok?
Test display math with equation numbers:
\begin{equation}
|\psi_1\rangle = a|0\rangle + b|1\rangle
\end{equation}
Is it ok?
Test display math with equation numbers:
$$
\begin{align}
|\psi_1\rangle &= a|0\rangle + b|1\rangle \\\\
|\psi_2\rangle &= c|0\rangle + d|1\rangle
\end{align}
$$
Is it ok?
Test display math with equation numbers:
\begin{align}
|\psi_1\rangle &= a|0\rangle + b|1\rangle \\\\
|\psi_2\rangle &= c|0\rangle + d|1\rangle
\end{align}
Is it ok?
March 29, 2016
Experimenting with R Markdown. The source is available here. We’ll use the iris dataset:
## Read in and summarize the data
data(iris)
str(iris)
## 'data.frame': 150 obs. of 5 variables:
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
## 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
## Median :5.800 Median :3.000 Median :4.350 Median :1.300
## Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
## 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
## Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
## Species
## setosa :50
## versicolor:50
## virginica :50
##
##
##
We see that there are 150 total observations of anatomical characteristics of three iris species. A visualization using ggplot2:
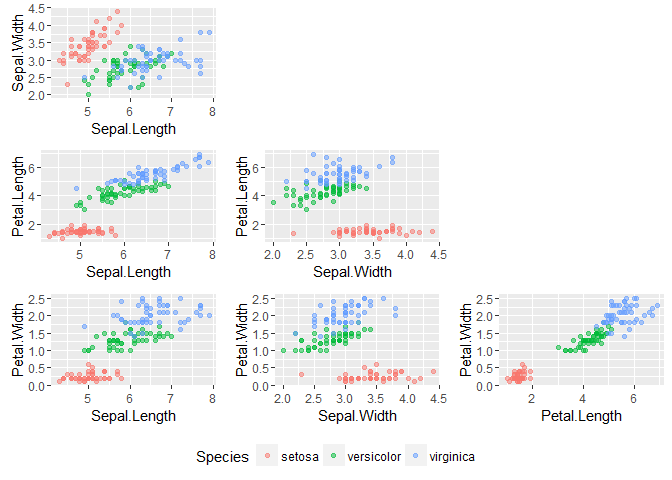
Observations:
- The setosa species is not like the others; it can be distinguished just on the basis of petal size
- It doesn’t look like versicolor and virginica can be completely separated
Try the linear discriminant as implemented in MASS.
library(MASS)
(iris.lda <- lda(Species ~ ., data=iris)) #enclosing in () makes R print the output
## Call:
## lda(Species ~ ., data = iris)
##
## Prior probabilities of groups:
## setosa versicolor virginica
## 0.3333333 0.3333333 0.3333333
##
## Group means:
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## setosa 5.006 3.428 1.462 0.246
## versicolor 5.936 2.770 4.260 1.326
## virginica 6.588 2.974 5.552 2.026
##
## Coefficients of linear discriminants:
## LD1 LD2
## Sepal.Length 0.8293776 0.02410215
## Sepal.Width 1.5344731 2.16452123
## Petal.Length -2.2012117 -0.93192121
## Petal.Width -2.8104603 2.83918785
##
## Proportion of trace:
## LD1 LD2
## 0.9912 0.0088
# Confusion matrix
table(Predicted=predict(iris.lda, iris[,1:4])$class,
Actual=iris$Species)
## Actual
## Predicted setosa versicolor virginica
## setosa 50 0 0
## versicolor 0 48 1
## virginica 0 2 49
Not half bad. But from the scatter plots we can get away with just petal length and width:
(iris.lda <- lda(Species ~ Petal.Length + Petal.Width, data=iris))
## Call:
## lda(Species ~ Petal.Length + Petal.Width, data = iris)
##
## Prior probabilities of groups:
## setosa versicolor virginica
## 0.3333333 0.3333333 0.3333333
##
## Group means:
## Petal.Length Petal.Width
## setosa 1.462 0.246
## versicolor 4.260 1.326
## virginica 5.552 2.026
##
## Coefficients of linear discriminants:
## LD1 LD2
## Petal.Length 1.544371 -2.161222
## Petal.Width 2.402394 5.042599
##
## Proportion of trace:
## LD1 LD2
## 0.9947 0.0053
# Confusion matrix
table(Predicted=predict(iris.lda, iris[,1:4])$class,
Actual=iris$Species)
## Actual
## Predicted setosa versicolor virginica
## setosa 50 0 0
## versicolor 0 48 4
## virginica 0 2 46
Not quite as clean, but cutting in half the number of features to be measured is (probably?) a win. Let’s visualize the (approximate) decision boundaries:
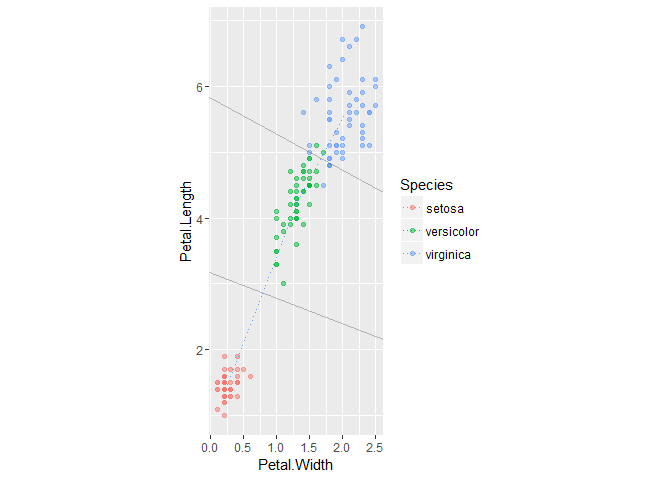
The darker points indicate overplotting, so the visual count of errors does line up with the confusion matrix above.
To show the actual boundaries I’d need to account for the relative covariances of the classes; not sure how to grab this from the model object but it can be done by hand. Doesn’t seem worthwhile for this example, though, so I’ll just trust MASS is doing its job, right?

